 Edmonds's Blossom Algorithm
Edmonds's Blossom Algorithm

Maximum matchings in general graphs
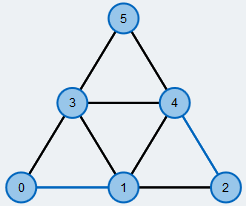
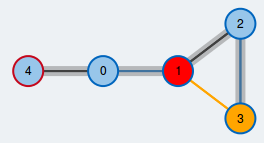
Maximum matching in a general graph.
Edges contained in the matching are colored blue.
An often occuring and well-studied problem in graph theory is finding a maximum matching in a graph \( G=(V,E)\). A matching M is a subset of edges such that every node is covered by at most one edge of the matching. M is a maximum matching if there is no other matching in G that has more edges than M.
This website is about Edmonds's Blossom Algorithm, an algorithm that computes a maximum matching in an undirected graph. In contrast to some other matching algorithms, the graph need not be bipartite. The algorithm was introduced by Jack Edmonds in 1965 and has been further improved since then. Many exact modern algorithms for the maximum matching problem on general graphs are still based on the ideas of the Blossom Algorithm.
Edmonds's Blossom Algorithm uses and extends the essential ideas of the Hopcroft-Karp algorithm, which computes a maximum matching for bipartite graphs. If you have not heard about this algorithm, we recommend having a look at it before proceeding with the Blossom Algorithm: Hopcroft-Karp Algorithm
We further assume that you are familiar with graph traversal, especially Breadth-First Search.
This website presents a visualization and detailed explanations of Edmonds's Blossom Algorithm. You can create a new graph, execute the algorithm on this graph to compute a maximum matching, and learn more about the theoretical backgrounds of the algorithm.
What do you want to do first?
Which graph do you want to execute the algorithm on?
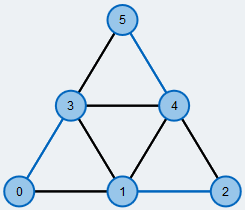
Start with an example graph:
Modify it to your desire:
- To create a node, make a double-click in the drawing area.
- To create an edge, first click on the output node and then click on the destination node.
- Right-clicking deletes edges and nodes.
Download the modified graph:
DownloadUpload an existing graph:
What next?
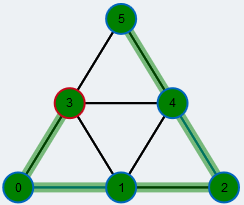
Legend
 |
yellow: free node |
 |
red stroke: root of the BFS |
 |
red: currently active node in BFS |
 |
orange: currently checked neighbor in BFS |
 |
green: node on the augmenting path |
 |
large radius: contracted node |
 |
orange: currently checked edge in BFS |
 |
blue: matched edge |
 |
grey overlay: edge in BFS tree |
 |
green overlay: edge on the augmenting path |
Legend
Algorithm status
Edmonds's Blossom Algorithm
The algorithm is ready to start now. Click "next" to execute a single step of the algorithm, "prev" to go one step back and "fast forward" for a fast execution of the algorithm.
Breadth-First Search finished
The BFS queue is empty. No augmenting path has been found by the modified Breadth-First Search.
Improved matching
The augmenting path is inverted to improve the matching. Inverting an augmenting path means that all unmatched edges along the path are changed to matched ones and vice versa. By doing so, the number of edges contained in the matching increases by 1. The new matching is colored blue.
Free vertices left
There are still unmatched vertices (yellow). We call them free vertices. We try to find an augmenting path to improve the matching. An augmenting path starts and ends at a free vertex and alternates between unmatched and matched edges.
Start Breadth-First Search
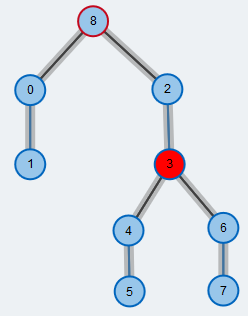
We pick one of the free vertices as the root node of a modified Breadth-First Search (BFS). Its stroke is colored red throughout the execution of the BFS. From the root node, we will construct a tree of alternating paths (BFS tree) until we reach another free vertex.
Pick next node from BFS queue
There are no neighbors left that we could check from the currently active node so we continue the BFS with the next node.
Contract blossom
The odd-length cycle we found is contracted to a single supernode. Supernodes are highlighted by a larger radius compared to the other nodes. We further push the new supernode to the BFS queue.
Continue BFS with next node
Pick the next node from the BFS queue. We call this node the currently active node and color it red. From this node, we will explore its neighbors to look for a free vertex.
Grow BFS tree
We add the neighbor and its mate to the BFS tree. The mate is further pushed to the BFS queue and we might continue our search from there later. The edges of the BFS tree are highlighted by a grey overlay.
Ignore even-length cycle
We can ignore even-length cycles. There is nothing to do in this case.
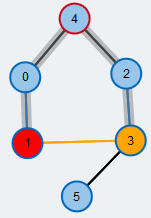
Check next neighbor
We check the next neighbor (orange) and find that it is already matched.
Check next neighbor
We check the next neighbor (orange edge) and find that it is a free vertex (yellow). Thus, there is an augmenting path between the root of the BFS and this vertex.
Check next neighbor
We check the next neighbor (orange) and find that this node is already contained in the BFS tree. Thus, there exists a cycle and we see that it has an odd number of edges. We call such a cycle a blossom.
Check next neighbor
We check the next neighbor (orange) and find that this node is already contained in the BFS tree. Thus, there exists a cycle and we see that it has an even number of edges.
Reconstruction of augmenting path
The augmenting path is highlighted green. Before improving the matching we have to expand the supernodes contained in the graph.
Expand supernodes
One of the supernodes is expanded and the augmenting path is adjusted correctly. There are still supernodes left that have to be expanded.
Reconstruction of augmenting path
The augmenting path from the BFS root to the free vertex found has been fully reconstructed and is highlighted green.
Reconstruction of augmenting path finished
After the expansion of all supernodes the augmenting path is fully reconstructed and highlighted green.
Algorithm finished
There are no free vertices in the graph anymore so we have found a matching of maximum cardinality. Its edges are colored blue.
BEGIN
WHILE F != ∅ DO (*set of free nodes F*)
pick r ∈ F
queue.push(r) (*BFS queue*)
T ← ∅ (*BFS tree T*)
T.add(r)
WHILE queue != ∅
v ← queue.pop()
FOR ALL neighbors w of v DO
IF w ∉ T AND w matched THEN
T.add(w)
T.add(mate(w))
queue.push(mate(w))
ELSE IF w ∈ T AND even-length
cycle detected THEN
CONTINUE
ELSE IF w ∈ T AND odd-length
cycle detected THEN
contract cycle
ELSE IF w ∈ F THEN
expand all contracted nodes
reconstruct augmenting path
invert augmenting path
END
If you switch tabs, the execution will be terminated.
You can open another browser window to read the description in parallel.